DNA Synthesis:
Take Home Message
Instructor: Dr. Natalia Tretyakova, Ph.D. «hyperlink
"mailto:[email protected]"»
- 6-3432
PDB reference
correction and design Dr.chem., Ph.D. Aris Kaksis, Associate Prof. e-mail:
[email protected]
1) DNA synthesis is
carried out by DNA polymerases with high
fidelity.
2)
DNA synthesis is characterized by initiation, priming and
processive synthesis steps and
proceeds in 5--> 3’ direction.
3) Modifications of
DNA base pairs, if not repaired, can
lead to mutations of the DNA sequence.
DNA
Replication
Long
before the structure of DNA was
known, scientists wondered at the ability of organisms to
create faithful copies of themselves and, later, at the ability of
cells to
produce many identical copies of large and complex macromolecules.
Speculation
about these problems centered around the concept of a template,
a structure
that would allow molecules to be lined up in a specific order and
joined, to
create a macromolecule with a unique sequence
and function. The 1940s brought the
revelation that DNA was the genetic
molecule, but not until James
Watson and Francis Crick deduced its structure
did it become clear how DNA could
act as a template for the replication
and transmission of genetic
information: one strand 1 is the
complement of the other 2 second The strict
base-pairing rules mean that each
strand provides the template for a sister strand with a predictable and complementary
sequence (see Figs 10-16, 10-17).
Besides maintaining the integrity
of DNA sequences by DNA repair, all
organisms must
duplicate their DNA accurately
before every cell division. DNA
replication occurs at polymerization
rates of about
500 nucleotides per second in bacteria
and about 50 nucleotides per second
in mammals. Clearly, the proteins that catalyze
this process must be both 2 accurate
and fast. Speed and accuracy
are
achieved by means of a multi-enzyme
complex that guides the process and constitutes an elaborate
"replication machine".
The fundamental properties of the
DNA replication process and the
mechanisms used by the enzymes that catalyze
it have proved to be
essentially identical in all organisms. This mechanistic unity is a
major theme
as we proceed from general properties of the replication
process, to E coli replication enzymes,
and,
finally, to replication in
eukaryotes.
DNA
Replication Is Governed by a Set of Fundamental Rules
DNA Replication Is Semi
Conservative.
Base-pairing Underlies DNA Replication
as well as DNA Repair
DNA templating is the
process in which the nucleotide sequence
of a DNA strand (or selected portions
of a DNA strand) is copied by complementary
base-pairing (A
with  or
or  , and G with C) into a complementary nucleic
acid sequence (either DNA
or RNA). The process entails the
recognition of each nucleotide in
the DNA strand by an unpolymerized complementary nucleotide and requires that the two 2 strands of the DNA helix be separated,
at least transiently, so that the hydrogen bond donor
and acceptor groups on each base become
exposed for base-pairing. The
appropriate incoming single 1
nucleotides are thereby aligned for their enzyme-catalyzed
polymerization into a new nucleic acid chain. In
1957 the first such nucleotide polymerizing enzyme, DNA polymerase, was discovered. The substrates
for this enzyme were found to be de-oxy-ribo-nucleoside
tri-phosphates,
which are polymerized on a single-stranded
DNA template. The stepwise mechanism of
this reaction is the one previously illustrated in DNA
Polymerase Enzyme in
connection with DNA repair. The
discovery of DNA polymerase led to
the isolation of RNA polymerase,
which was correctly
inferred to use ribo-nucleoside
tri-phosphates as its substrates.
, and G with C) into a complementary nucleic
acid sequence (either DNA
or RNA). The process entails the
recognition of each nucleotide in
the DNA strand by an unpolymerized complementary nucleotide and requires that the two 2 strands of the DNA helix be separated,
at least transiently, so that the hydrogen bond donor
and acceptor groups on each base become
exposed for base-pairing. The
appropriate incoming single 1
nucleotides are thereby aligned for their enzyme-catalyzed
polymerization into a new nucleic acid chain. In
1957 the first such nucleotide polymerizing enzyme, DNA polymerase, was discovered. The substrates
for this enzyme were found to be de-oxy-ribo-nucleoside
tri-phosphates,
which are polymerized on a single-stranded
DNA template. The stepwise mechanism of
this reaction is the one previously illustrated in DNA
Polymerase Enzyme in
connection with DNA repair. The
discovery of DNA polymerase led to
the isolation of RNA polymerase,
which was correctly
inferred to use ribo-nucleoside
tri-phosphates as its substrates.
During DNA replication each of the two 2
old DNA strands serves
as a template for the formation of
an entire new strand. Because each
of the two 2 daughters of a dividing
cell inherits a new DNA double
helix containing one 1 old
and one 1 new strand (see Figure
25-2), DNA
is said to be replicated "semi
conservatively" by DNA polymerase.
Each DNA strand serves as a template
for the synthesis of a new strand,
producing two new DNA molecules, each with one
new strand and one old strand. This is
semi conservative replication.
The hypothesis of semi conservative replication was
proposed by Watson and Crick soon after publication of their 1953 paper
on the structure of DNA, and was
proved by ingeniously designed experiments carried out
by Matthew Meselson and Franklin Stahi in 1957. Meselson and Stahi grew
E.
coli cells for many generations in a medium in which the sole nitrogen N
source (NH4Cl)
contained 15N, the "heavy"
isotope of nitrogen N,
instead of the normal,
more abundant "light"
isotope, 14N. The DNA
isolated from these cells had a density about 1%
greater than that of normal
['14N] DNA
(Fig. 25-2a). Although this is only a small difference, a mixture of
heavy [15N] DNA
and light [14N]DNA can be separated
by centrifugation to equilibrium in a
cesium chloride CsCl density
gradient.
The E. coli cells grown in
the 15N medium were transferred to a fresh
medium
containing only the 14N isotope, where they were
allowed to grow until the cell population had just doubled. The DNA isolated from these first-generation
cells formed a single 1 band
in the CsCl
gradient at a position indicating that the
double-helical DNAs of the daughter
cells were hybrids containing one 1 new 14N strand and
one 1 parental 15N strand
(Fig. 25-2b).
This result argued against conservative replication,
an
alternative hypothesis in which one 1
progeny DNA molecule would consist of two 2 newly
synthesized DNA strands and the other would contain the
two 2 parental strands; this would
not yield hybrid DNA molecules in
the Meselson-Stahl experiment. The semi
conservative replication hypothesis was further supported in the next
step of the experiment (Fig. 25-2c).
Cells were again allowed to double 2
in number in the 14N medium. The isolated DNA
product of this second 2
cycle of replication exhibited two 2 bands
in the density gradient, one with a
density equal to that of light DNA
and the other with the density of the hybrid
DNA observed after the first cell doubling.
DNA extracted and centrifuged to equilibrium in CsCl density gradient













Figure 25-2. The Meselson-Stahl experiment.
(a) Cells were grown for many generations
in a medium containing only heavy nitrogen, 15N, so that
all the nitrogen
N in their DNA was 15N, as shown
by a single 1 band (blue)
when centrifuged in
a
CsCl
density gradient.
(b) Once the cells had been
transferred to a medium containing only light nitrogen, 14N, cellular DNA
isolated after one generation
equilibrated at a higher position in the density gradient (purple band).
(c) Continuation of replication for a
second 2 generation yielded two 2 hybrid
DNAs and two 2 light
DNAs (red), confirming semi
conservative replication.
Possible
Models for DNA Replication







Conservative replication Dispersive
replication
Semiconservative
replication












Possible
Models os DNA Replication
 Meselson,
Stahl 1958
Meselson,
Stahl 1958
DNA
Polymerization
(DNA)n bases + dNTP -->
(DNA)(n+1) bases + PPi

E. coli DNA
Polymerase I
Klenow
Fragment
N-

 –C
–C
36 kDa
67 kDa
•
large cleft for duplex
DNA
•
flexible finger and
thumb region for positioning of duplex and dNTPs polymerase site
•
3'-->
5'
and 5'--> 3'
exonuclease catalytic sites
3` --> 5` Exonuclease
5`--> 3` Exonuclease

Typical Polymerase Structure

KLENOW
Polymerase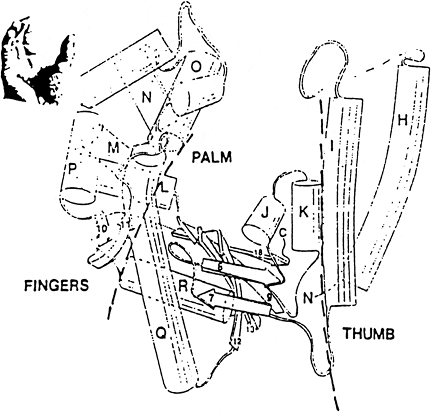
Typical
Polymerase Structure

Exonuclease
Domain

![]() or
or ![]() , and G with C) into a complementary nucleic
acid sequence (either DNA
or RNA). The process entails the
recognition of each nucleotide in
the DNA strand by an unpolymerized complementary nucleotide and requires that the two 2 strands of the DNA helix be separated,
at least transiently, so that the hydrogen bond donor
and acceptor groups on each base become
exposed for base-pairing. The
appropriate incoming single 1
nucleotides are thereby aligned for their enzyme-catalyzed
polymerization into a new nucleic acid chain. In
1957 the first such nucleotide polymerizing enzyme, DNA polymerase, was discovered. The substrates
for this enzyme were found to be de-oxy-ribo-nucleoside
tri-phosphates,
which are polymerized on a single-stranded
DNA template. The stepwise mechanism of
this reaction is the one previously illustrated in DNA
Polymerase Enzyme in
connection with DNA repair. The
discovery of DNA polymerase led to
the isolation of RNA polymerase,
which was correctly
inferred to use ribo-nucleoside
tri-phosphates as its substrates.
, and G with C) into a complementary nucleic
acid sequence (either DNA
or RNA). The process entails the
recognition of each nucleotide in
the DNA strand by an unpolymerized complementary nucleotide and requires that the two 2 strands of the DNA helix be separated,
at least transiently, so that the hydrogen bond donor
and acceptor groups on each base become
exposed for base-pairing. The
appropriate incoming single 1
nucleotides are thereby aligned for their enzyme-catalyzed
polymerization into a new nucleic acid chain. In
1957 the first such nucleotide polymerizing enzyme, DNA polymerase, was discovered. The substrates
for this enzyme were found to be de-oxy-ribo-nucleoside
tri-phosphates,
which are polymerized on a single-stranded
DNA template. The stepwise mechanism of
this reaction is the one previously illustrated in DNA
Polymerase Enzyme in
connection with DNA repair. The
discovery of DNA polymerase led to
the isolation of RNA polymerase,
which was correctly
inferred to use ribo-nucleoside
tri-phosphates as its substrates.


 Meselson,
Stahl 1958
Meselson,
Stahl 1958





