Instructor: Dr. Natalia Tretyakova, Ph.D.
«hyperlink
"mailto:[email protected]"»
- 6-3432
PDB reference correction and design Dr.chem.,
Ph.D. Aris
Kaksis, Associate Prof. mailto:[email protected]
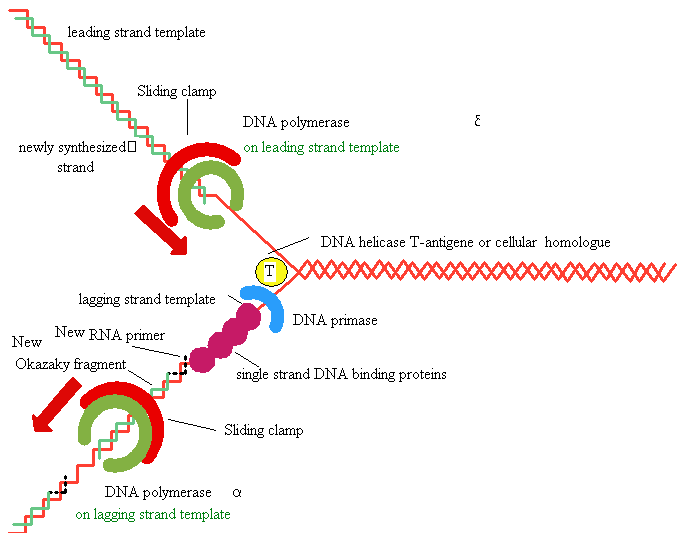
5'
3' DNA SynthesisDNA Synthesis

3'
5'
3'
5'
Primer
Synthesis and removal
3'  5'
5'
¯
Primase
3'  5'
5'
5'  3'
3'
RNA primer
 + ¯ DNA Pol
III
+ ¯ DNA Pol
III
3'  5'
5'
5' 
 ¾® 3'
¾® 3'
¯ DNA Pol I
DNA strand
3'
 5'
5'
5'  3'
3'
degraded 






 ¬ primer + ¯ DNA
Ligase
¬ primer + ¯ DNA
Ligase
3'  5'
5'
5'  3'
3'
Replication Fork Garland



Fidelity
of Polymerization: Absolutely Essential!!
Error Probability =
Polymerization error 10-4
3' --> 5' Nuclease error 10-3
(10-4) •
(10-3)
= 10-7 or 1 in
10,000,000
DNA
Synthesis: addition of
new dNTPs follows Watson-Crick rules
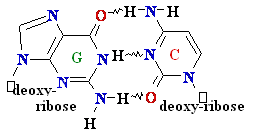
G =
C
A=
Polymerase
errors
• Very low rate of
misincorporation (1 per 108)
• Errors
can occur due to the presence of minor
tautomers of nucleobases.
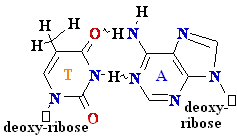
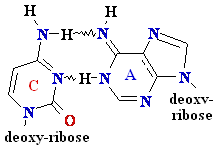
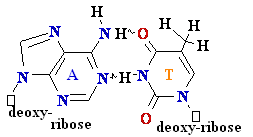
 = A
Cytosine
C = A Rare tautomer of Adenine
= A
Cytosine
C = A Rare tautomer of Adenine
Normal base pairing
Mispairing
Proofreading
function of DNA polymerases


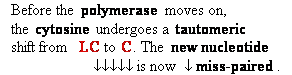



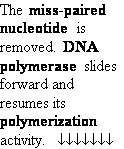
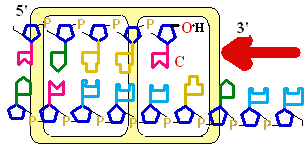
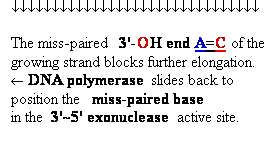
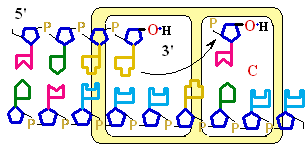
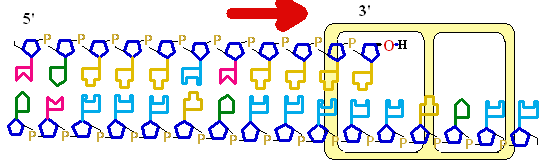
Figure 25-7. An
example of error correction by the 3'--> 5'
exonuclease activity
of DNA polymerase I.
Structural analysis has located the exonuclease
activity ahead of the polymerase
activity as the enzyme is oriented
in its movement along the DNA. A miss-matched
base (here, a C=A mismatch) impedes
translocation of DNA polymerase I
to the next site. Sliding backward, the enzyme
corrects the mistake with its 5'--> 3' exonuclease
activity, then resumes its polymerase
activity in the 5'--> 3'
direction.
Consider
misincorporation due to a rare tautomer of A
2nd replication -A-
- -
-
1st replication -A(imino)-
|
-C-
5’-A- --> -A(imino) |
3’- - --> -
- --> - -
|
-
|
-A-
- - Normal
replication
- Normal
replication
Final result: A -->
G
transition
Mismatch Repair Enzymes
Polymerase I, III error rates: 1 per
107
nucleotides
Observed mutation rate:
1
per 108 - 1 per 1010 nucleotides
Polymerase errors
can be corrected after DNA synthesis!
Repair of nucleotide mismatches:
• Recognize parental
DNA strand (correct base) and daughter
strand (incorrect base)
Parental strand is methylated —CH3: metC 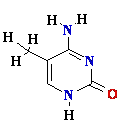 or
or 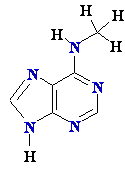 Amet
Amet
2. Replace
a portion of the strand containing erroneous
nucleotide
(between the mismatch and
a nearby methylated
site –up to 1000 nt)
DNA
replication in eukaryotes
Several eukaryotic
DNA polymerases are known: a, b,
d,
g -
a
and d are thought to be
the major chromosomal replicases
Similarities with E.Coli
Always 5’
to 3’ direction -->
Require a primer
Similarities in active site and tertiary 3° structure
Differences
Eukaryotic replication is
much slower (100
nt/sec)
Several replication
origins
Polymerases are more
specialized (a for lagging strand, d for leading strand)
4. Require special
processing of the chromosomal ends .
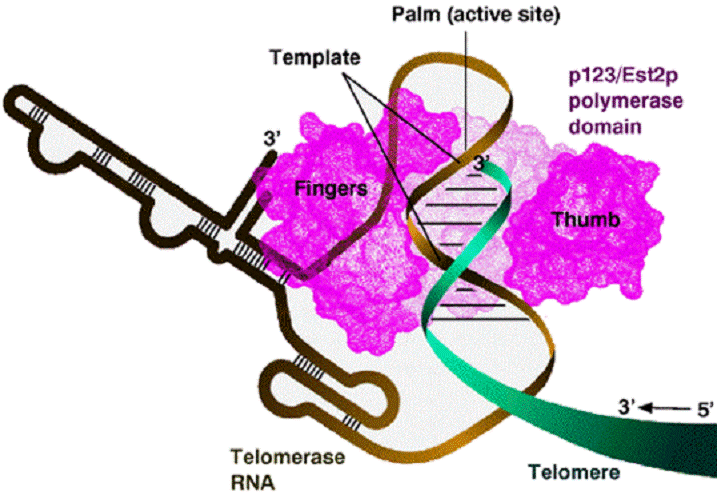
Telomerase preserves
chromosomal ends
• The ends of the linear DNA strand
can not be replicated due to the lack
of a primer
• This would lead to shortening
of DNA strands after
replication
3'  5'
5'
5' 
 3'
<--RNA
primer
3'
<--RNA
primer
• Solution: the chromosomal
ends are extended by DNA telomerase
This enzyme adds hundreds 200÷900
of tandem repeats of a hexa-nucleotide(AGGG
 in
humans)
in
humans)
to
the parental strand:
3'
 5' AGGG
5' AGGG
 AGGG
AGGG
 AGGG
AGGG
 <-- telomere
<-- telomere
5' 

 3'
||||||||||||¯
3'
||||||||||||¯
3'
 5'
AGGG
5'
AGGG
 AGGG
AGGG
 AGGG
AGGG
 <-- telomere
<-- telomere
5'  3'
3'  CCCAA
CCCAA CCCAA
CCCAA CCCAA<-- RNA primer
CCCAA<-- RNA primer
Telomerase is a
ribonucleoprotein that contains an RNA molecule
used as a template for elongation of the 3’ strand
DNA ||-------------->
RNA --------------> Proteins ----->>>>>>> Cellular action
Replication || || transcription
||||||| translation
------>>>>||||||||||||
|| nucleare cytosolic
DNA
nucleare <= Reverse
transcription
of telomeres
Notable
exception: retroviruses
RNA ||------------>
DNA -------->
RNA ------------------> Proteins ----------> Cellular action
Reverse ||->||transcription transcription
||||||| translation
--->>>
||||||||||||
DNA cytosolic
nucleare
cytosolic
Reverse
transcriptases (RT)
are RNA
directed DNA Pol
Used by RNA viruses (HIV-I
, human immunoblastosis
virus, Rous sarcoma virus) :
1. Make RNA-DNA hybrid (use its own
RNA as a primer)
2. Make ss DNA by exoribonuclease (RNase H)
activity
3. Make ds DNA incorporate
in the
host genome
 CCCAA
CCCAA CCCAA
CCCAA CCCAA RNA
CCCAA RNA
||<--- RT
 CCCAA
CCCAA CCCAA
CCCAA CCCAA RNA
CCCAA RNA
AGGG
 AGGG
AGGG
 AGGG
AGGG
 - DNA hybrid
- DNA hybrid
||<--- Rnase H --------------------->  CCCAA
CCCAA CCCAA
CCCAA CCCAA
RNA
CCCAA
RNA
||<--- RT
AGGG
 AGGG
AGGG
 AGGG
AGGG
 ss -DNA
ss -DNA
||<--- RT
 CCCAA
CCCAA CCCAA
CCCAA CCCAA
ds
DNA
CCCAA
ds
DNA
AGGG
 AGGG
AGGG
 AGGG
AGGG
 ds -DNA
ds -DNA
Termination
of
Polymerization: The Key to Nucleoside Drugs

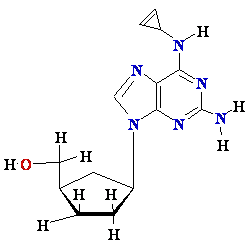
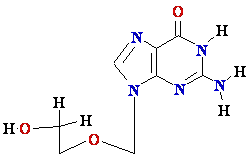
AZT
Ziagen
Acyclovir
Inhibition of
Viral DNA Polymerization by nucleoside analogs
(DNA)n bases + dNTZiagen (DNA)(n+1) bases analog ¹ 

 E. coli DNA
Polymerase I
E. coli DNA
Polymerase I
Nucleosides Must Be Converted
to Triphosohates to be
Part of DNA and RNA
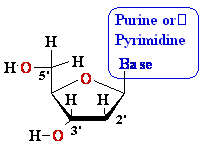

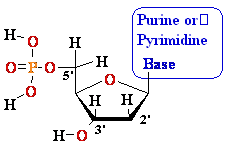

MonoPhosphate
¯
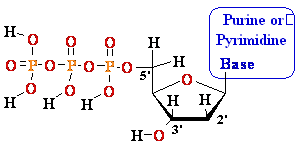

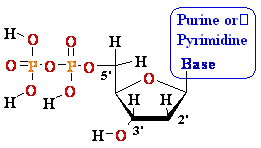
TriPhosphate
DiPhosphate
Chemical
modification of DNA
Carcinogen (X) -------------------> detoxification ----> excretion
metabolic activation || 
reactive metabolite (X-)
+ DNA
 ||
DNA adducts
||
DNA adducts
 || repair
||
|| replication
|| repair
||
|| replication
intact DNA
cell death mutations

Types
of DNA Mutations
1.
Point mutations:
substitution of one base
pair for another, e.g. A for GC
for GC
• the most common form of mutation
• transition;
purine to purine and
pyrimidine to pyrimidine
•transversions; purine to pyrimidine or
pyrimidine to purine
2.
Deletion of one or
more base pairs
3.
Insertion of
one or more base pairs
DNA
Damage
Sources of DNA
damage: endogenous
1.
Deamination
2.
Depurination: 10,000/cell/day
3.
Oxidative stress
Sources of DNA
damage: environmental
1. Alkylating agents (drugs, pollutants)
2. X-ray and UV irradiation
3. Diet
4. Smoking
Mechanisms of
induced mutations
·
Altered basepairing characteristics (O6-alkyl-G)
·
Abasic sites (N7-guanine
adducts)
·
Deletions/insertions due to
intercalating agents
(e.g acridin orange)
·
DNA strand breaks
(reactive oxygen species)
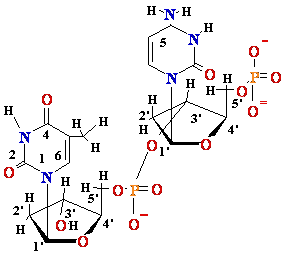
Normal base
pairing in DNA and
an example of mispairing via chemically modified nucleobase
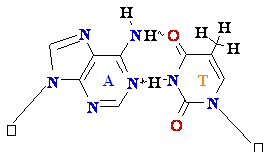

Adenine A=
 O6-Alkyl-Guuanine
O6-Alkyl-Guuanine 
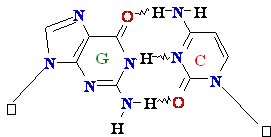
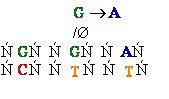
Guanine
GºC
Cytosine
DNA Damage:
deamination
 -->
-->  Adenine
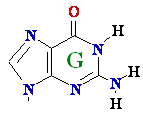
A --> hypoXanthine
Adenine
A --> hypoXanthine
 ->
->  Guanine
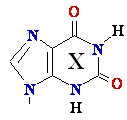
G --> Xanthine
Guanine
G --> Xanthine
 C
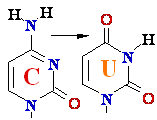
Cytosine -->
C
Cytosine --> 

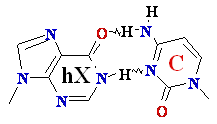
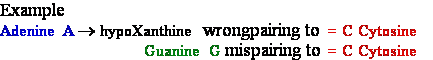


 C Cytosine Deamination to ®
C Cytosine Deamination to ® 

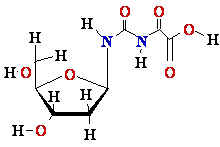
G Guanine
Depurination G or
A remove by
hydrolise H2O
 ¾®
¾® Abasic site
Abasic site
DNA Damage: oxidative stress
Reactive
oxygen species:
HO•, H2O2, 1O2, LOOH

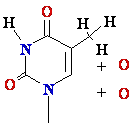

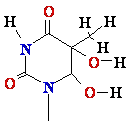
 glycol
glycol
Guanine


 8-oxo-Guanine
8-oxo-Guanine
Guanine
oxidation in DNA
LOO•
HO•, HOCl, 1O2, HONO LOO• HO•,
HOCl, 1O2, HONO
||
||
||
||
|| ||
||
||
 ----->
----->
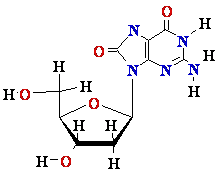
dG in DNA ||
8-oxo-dG in DNA
||
||
||
||
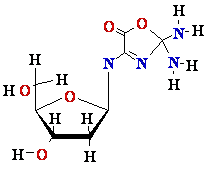

DNA Damage: UV
light


Also
 = C
, C = C dimers-neighbour
= C
, C = C dimers-neighbour
 dimer-neighbour
dimer-neighbour
 =
=
Benzo[a]pyrene-
induced DNA
adducts
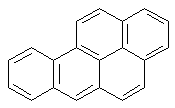 ----->
----->
benzo[a]pyrene
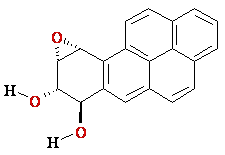
(+)-trans-anti
BPDE |reaction|
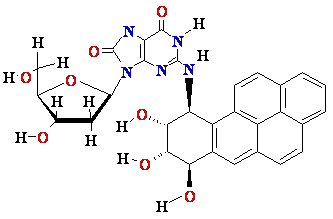
N2-BPDE-dG
adduct
Structure of N2-BPDE-dG
containing DNA



![]() 5'
5' ![]() 5'
5' ![]() 3'
3' ![]() + ¯ DNA Pol
III
+ ¯ DNA Pol
III ![]() 5'
5' ![]()
![]() ¾® 3'
¾® 3'
![]() 5'
5' ![]() 3'
3' ![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]() ¬ primer + ¯ DNA
Ligase
¬ primer + ¯ DNA
Ligase ![]() 5'
5' ![]() 3'
3' 





![]()


![]() = A
Cytosine
C = A Rare tautomer of Adenine
= A
Cytosine
C = A Rare tautomer of Adenine 

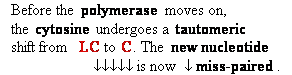



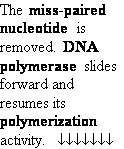

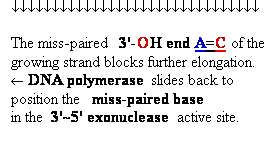


![]() -
-![]() - --> -
- --> -![]() -
|
-
| ![]() -
-  or
or  Amet
Amet




 E. coli DNA
Polymerase I
E. coli DNA
Polymerase I










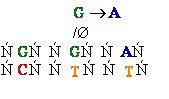
 -->
-->  Adenine
A --> hypoXanthine
Adenine
A --> hypoXanthine
 ->
->  Guanine
G --> Xanthine
Guanine
G --> Xanthine
 C
Cytosine -->
C
Cytosine --> 
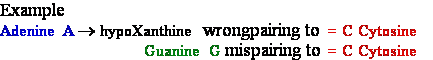


 C Cytosine Deamination to ®
C Cytosine Deamination to ® 
 ¾®
¾® Abasic site
Abasic site





 8-oxo-Guanine
8-oxo-Guanine ----->
----->







 ----->
----->



